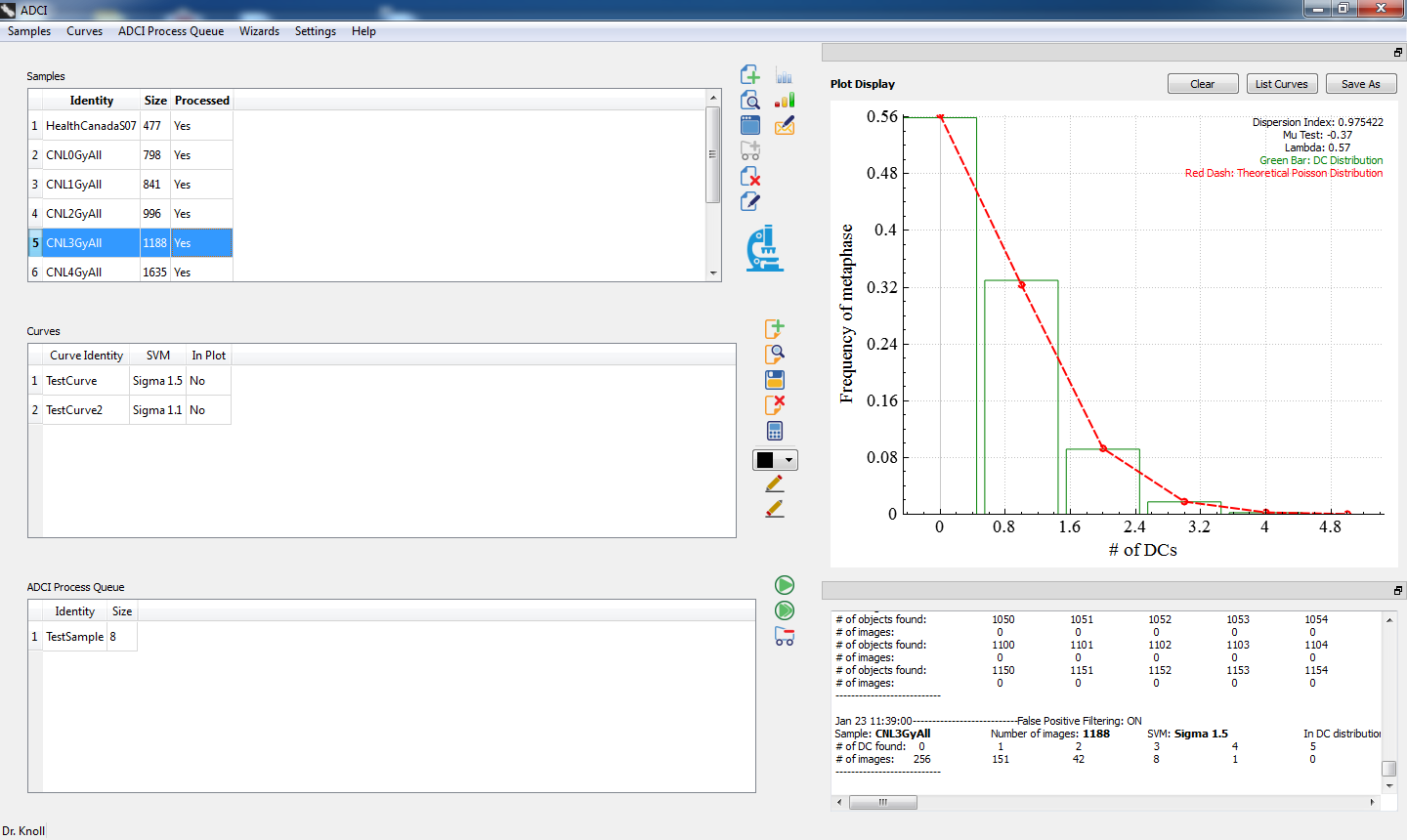
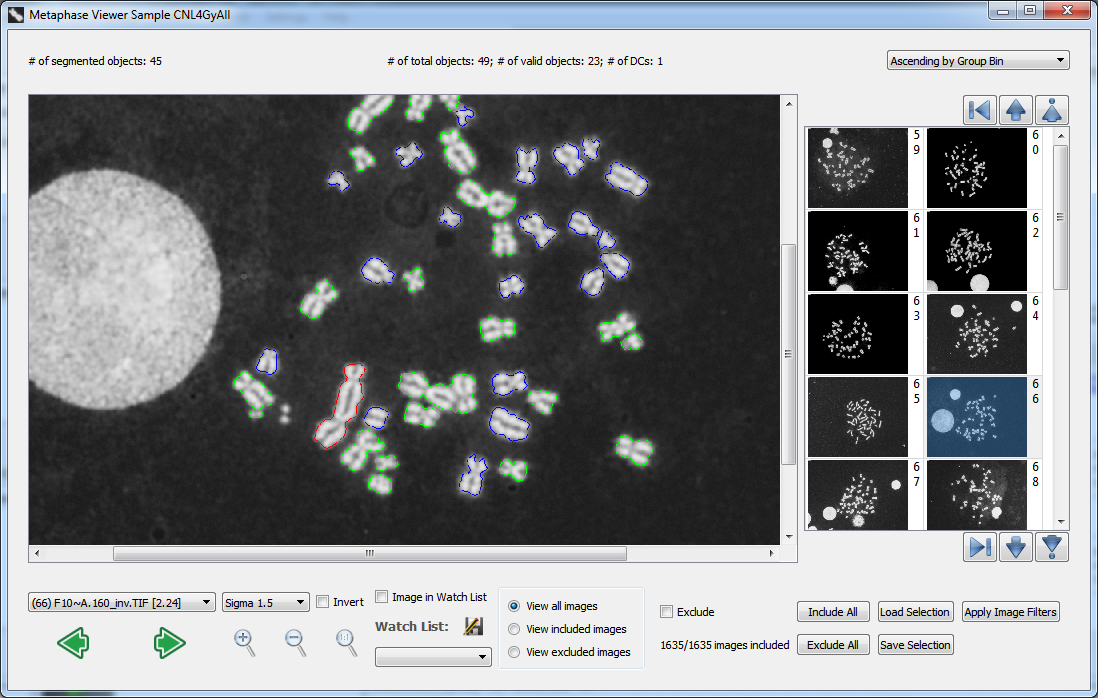
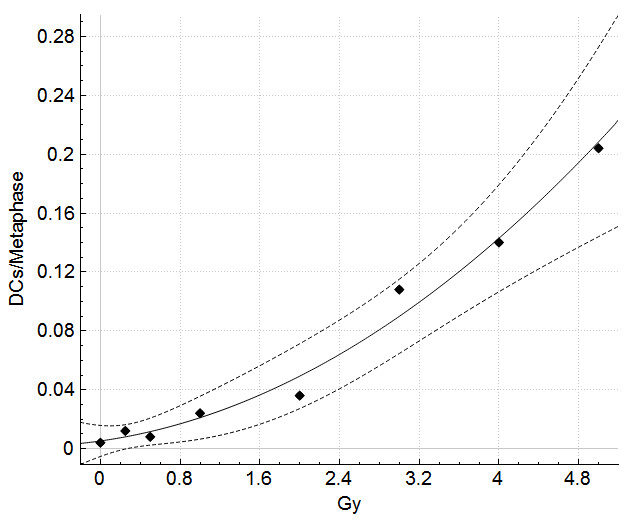
A straightforward single-screen interface provides access to all samples, curves, plots, console, and one-click sample processing.
overview
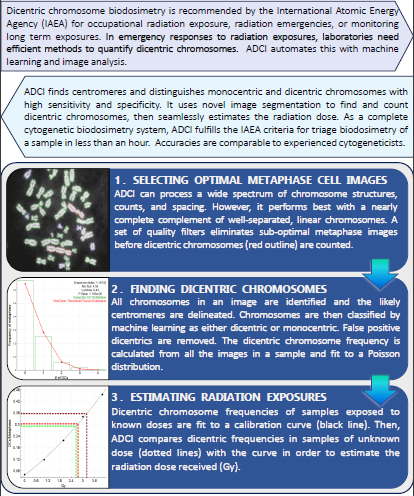
Automated Dicentric Chromosome Identifier and Radiation Dose Estimator (ADCI) software was developed by CytoGnomix Inc. to rapidly estimate radiation doses of multiple exposed individuals. ADCI is available as desktop software for Microsoft Windows or as a cloud-based subscription service (ADCI Online overview) streamed through a web browser. The dicentric chromosome assay is the gold standard for assessing biological radiation exposure (IAEA and WHO). Labor-intense manual analysis of dicentric chromosomes in 100's to 1000's of metaphase images by trained personnel is no longer required. ADCI selects images, detects dicentric chromosomes quickly (~3 min/sample), computes dicentric frequencies, creates calibration curves, and estimates dose.
More information about all ADCI functionality can be found within our online wiki. For a brief introduction to the ADCI's main workflow and user interface consult our one page shortcut document. To learn more about the underlying algorithm employed by ADCI to perform dicentric chromosome detection or to read about the performance of ADCI on datasets supplied to us, view the publication history of ADCI. View a 10 minute video available in the Journal of Visualized Experiments (JoVE) for a brief demonstration of the software. Please contact us with any additional queries. Visit our frequently asked questions page to read answers to questions we’ve previously received.
A demonstration version of ADCI is available at no cost. Click here to request a download link. A quote for the licensed version of ADCI can be requested here.
We have also developed gene expression signatures based on machine learning of radiation responsive genes. Please see:
- Zhao JZL, Mucaki EJ and Rogan PK. Predicting ionizing radiation exposure using biochemically-inspired genomic machine learning [version 2; referees: 3 approved]. F1000Research 2018, 7:233 (doi: 10.12688/f1000research.14048.2).
- Eliseos J. Mucaki, Ben C. Shirley & Peter K. Rogan (2021) Improved radiation expression profiling in blood by sequential application of sensitive and specific gene signatures, International Journal of Radiation Biology (doi: 10.1080/09553002.2021.1998709).
adci online new
ADCI_Online is a subscription service which grants access to ADCI on a dedicated cloud-based system through a web browser. Interacting with ADCI “on the cloud”:
- Eliminates the need for a dedicated computer system to run ADCI
- Generates the same results as dedicated systems
- Has a short-term subscription format which reduces cost
- Can accommodate on-demand bursts of computing power when necessary
- Can ensure that cytogenetic data is colocated in the same region as the user
- Securely isolates individual user data and protects software analysis from intrusion, disruption and corruption
- Allows dose estimation to be carried out anywhere there is a reliable internet connection
Although each system running ADCI_Online has less computing power than a high-performance system running ADCI Desktop, the cloud-based nature of ADCI_Online allows for rapid expansion of resources. The system can be cloned as many times as necessary, resulting in an array of cloud-based systems available for use.
Metaphase images and ADCI_Online output are stored in an encrypted S3 bucket (Amazon Web Services). Additionally, files are encrypted while in transit to/from the bucket.
Documentation related to ADCI_Online can now be found within our online wiki.
Please contact CytoGnomix if you have additional questions about ADCI_Online.
features
Extensive documentation covering these and all other features of ADCI software can be found in our online documentation.
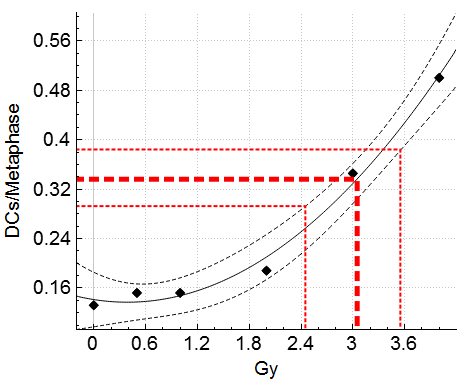
Dose estimate calculations can take into account uncertainty due to the calibration curve and uncertainty due to the Poisson nature of dicentric yield.
software demonstration
This 10 minute video published in the Journal of Visualized Experiments demonstrates how ADCI can be used to estimate the dose (in Gy) of test samples exposed to an unknown dose of radiation. Steps within the software necessary to do so, such as generation of calibration curves and selection of images, are described and demonstrated. The full text of the article provides additional details. Note there is an option to expand the video to a full screen view after the play button is clicked.
testimonials
I have assessed ADCI v1.9 and find its speed and accuracy is suitable for emergency response biodosimetry. Health Canada will be adding it to our toolkit for managing performing dose estimates after exposures from large scale radiological or nuclear events.
— Ruth Wilkins, Director, Radiobiology Laboratory, Consumer and Clinical Radiation Protection Bureau, Health Canada
ADCI is an important development for radiation dose estimation. It is essential that we advocate for its adoption to improve radiation safety internationally.
— Edward Waller, NSERC Industrial Research Chair, Faculty of Energy Systems and Nuclear Science, Ontario Institute of Technology
Thank you again for coming here and demonstrating and delivering the software, it was beneficial to all of us.
— Farrah Flegal, Director, Biodosimetry Laboratory, Canadian Nuclear Laboratories
This product is a game changer in the biodosimetry field.
— Ruth Wilkins, Director, Radiobiology Laboratory, Consumer and Clinical Radiation Protection Bureau, Health Canada
I think ADCI will be better than DCscore software (Metasystems) because you have integrated many image selection models. By this way, the dicentric detection ability of your system will be more effective.
— Pham Ngoc Duy, Biodosimetry Section, Center for Biotechnology, Nuclear Research Institute, Vietnam
This paper [F1000Research article] presents a considerable amount of work devoted to improving the accuracy of existing automatic dicentric systems for biological dosimetry. The authors have developed some very interesting ideas in constructing filters that detect false positive dicentrics and thereby improve the accuracy of 'hands-off' microscopy. The work has demonstrated that inter-laboratory differences, commonly reported in biological dosimetry, and evident between the two labs here, can be considerably improved by application of the filters. It is particularly interesting to see how the linear dose response reported by one lab, over a dose range that should show a lin/quadratic curve, can indeed be converted to linear quadratic by filtering out the false positives. Moreover they have demonstrated that much of the improvement can be achieved by a sub-set of their filters which should simplify future developments by not needing to employ all the methods.
The single most pressing remaining problem is the selection of metaphases of sufficient quality for passing onto the filtration procedures. Manual selection still seems better for removing the wide range of unsuitable material although the authors have demonstrated that the automated approach is getting there. ... I agree that fully automated image selection would be particularly advantageous when rapid triage dosimetry sorting of many cases is needed.
It is gratifying to see that the accuracy of dose estimations using the procedures described here falls well within the requirements for triage sorting following a major radiological incident.
Overall they are to be congratulated on a well-presented account of an improved approach to automated ‘dicentric-hunting’.
— David Lloyd, Centre for Radiation, Chemical and Environmental Hazards (CRCE), Public Health England, Didcot, UK
This paper [F1000Research article] presents an approach for the improvement of automatic detection of dicentrics and particularly the removal of some kinds of False Positives (FP) ... the manual scoring of dicentric chromosome is time consuming and the improvement of the result time is necessary in biological dosimetry. For dose assessment in case of triage of radiation exposed population, automatization of dicentric detection is very useful.
In order to increase the dicentric detection, the authors concentrate the study on the removal of FPs as well as on the selection of quality images. Apparently the main FPs in the metaphases of the 2 study laboratories are Sister Chromatid Separation (SCS) (84% of the FPs). This study shows that the authors reach to eliminate them with filters (about 55% of SCS removed). This is great and encouraging to remove other kind of FPs.
The study showed also the importance of selecting metaphases before the analysis and the detection. The quality of images and chromosomes is an important factor. The dose assessment becomes better when the images were selected than when they are not selected. The data show a difference in dose assessment between the 2 labs. The filters gave better results for CNL images than for HC images particularly on low doses.
However the selection of good quality metaphases is also time-consuming. There is a good comparison between manual and automatic triage of metaphases for the 2 labs highlighted by the statistical tests.
... Overall the authors are to be congratulated on a well-presented study of their work.
Eric Gregoire, Biological Dosimetry Laboratory (LDB), Institute for Radiological Protection and Nuclear Safety (IRSN), Fontenay-aux-Roses, France